Describe the experimental rationale that allowed the lac repressor to be isolated.

Erythritol, a natural sugar abundant in fruits and fermenting foods, is about 65 percent as sweet as table sugar and has about 95 percent fewer calories. It is 'tooth friendly' and generally devoid of negative side effects as a human consumable product. Pathogenic Brucella strains that catabolize erythritol contain four closely spaced genes, all involved in erythritol metabolism. One of the four genes (eryD) encodes a product that represses the expression of the other three genes. Erythritol catabolism is stimulated by erythritol. Present a simple regulatory model to account for the regulation of erythritol catabolism in Brucella. Does this system appear to be under inducible or repressible control?
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Gene Regulation in Prokaryotes
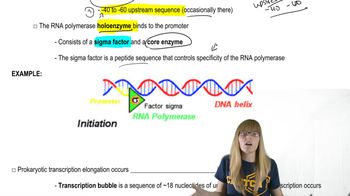
Inducible vs. Repressible Operons
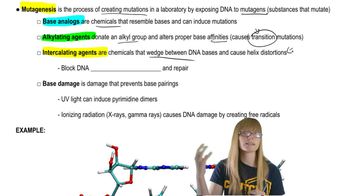
Negative Feedback and Repressor Proteins

What properties demonstrate that the lac repressor is a protein? Describe the evidence that it indeed serves as a repressor within the operon system.
Predict the effect on the inducibility of the lac operon of a mutation that disrupts the function of:
(a) The CRP gene, which encodes the CAP protein
(b) The CAP-binding site within the promoter.
Describe the role of attenuation in the regulation of tryptophan biosynthesis.
Neelaredoxin is a 15-kDa protein that is a gene product common in anaerobic bacteria. It has superoxide-scavenging activity, and it is constitutively expressed. In addition, its expression is not further induced during its exposure to O₂ or H₂O₂ [Silva, G. et al. (2001). J. Bacteriol. 183:4413 4420]. What do the terms constitutively expressed and induced mean in terms of neelaredoxin synthesis?
