In recombination studies of the rII locus in phage T4, what is the significance of the value determined by calculating phage growth in the K12 versus the B strains of E. coli following simultaneous infection in E. coli B? Which value is always greater?

 Klug 12th Edition
Klug 12th Edition Ch. 6 - Genetic Analysis and Mapping in Bacteria and Bacteriophages
Ch. 6 - Genetic Analysis and Mapping in Bacteria and Bacteriophages Problem 20a
Problem 20aUsing mutants 2 and 3 from Problem 19, following mixed infection on E. coli B, progeny viruses were plated in a series of dilutions on both E. coli B and K12 with the following results.
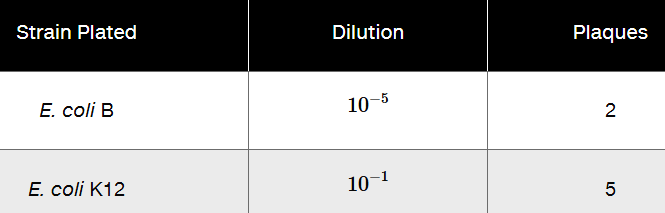
What is the recombination frequency between the two mutants?
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Recombination Frequency

Mutants in Genetics

Plaque Assay

In an analysis of rII mutants, complementation testing yielded the following results:
If further testing of the mutations in Problem 18 yielded the following results, what would you conclude about mutant 5?
Using mutants 2 and 3 from Problem 19, following mixed infection on E. coli B, progeny viruses were plated in a series of dilutions on both E. coli B and K12 with the following results.
Another mutation, 6, was tested in relation to mutations 1 through 5 from Problems 18–20. In initial testing, mutant 6 complemented mutants 2 and 3. In recombination testing with 1, 4, and 5, mutant 6 yielded recombinants with 1 and 5, but not with 4. What can you conclude about mutation 6?
During the analysis of seven rII mutations in phage T4, mutants 1, 2, and 6 were in cistron A, while mutants 3, 4, and 5 were in cistron B. Of these, mutant 4 was a deletion overlapping mutant 5. The remainder were point mutations. Nothing was known about mutant 7. Predict the results of complementation (+ or -) between 1 and 2; 1 and 3; 2 and 4; and 4 and 5.