In analyzing genetic data, how do we know whether deviation from the expected ratio is due to chance rather than to another, independent factor?
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
3. Extensions to Mendelian Inheritance
Chi Square Analysis
Problem 13c
Textbook Question
Researchers cross a corn plant that is pure-breeding for the dominant traits colored aleurone (C1), full kernel (Sh), and waxy endosperm (Wx) to a pure-breeding plant with the recessive traits colorless aleurone (c1), shrunken kernel (sh), and starchy (wx). The resulting F₁ plants were crossed to pure-breeding colorless, shrunken, starchy plants. Counting the kernels from about 30 ears of corn yields the following data.
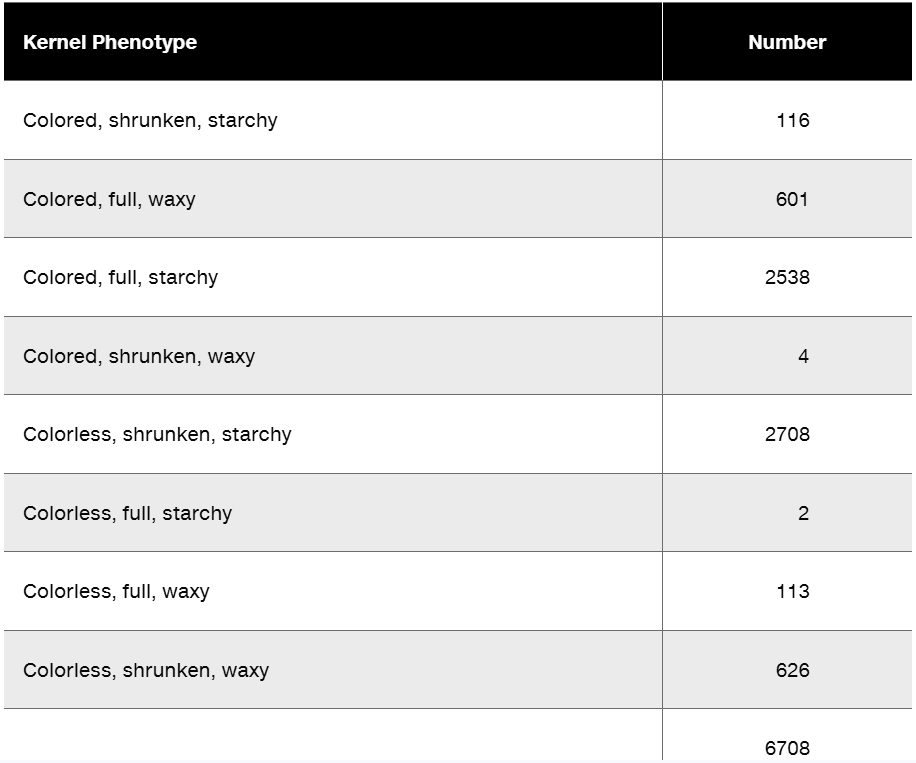
What is the order of these genes in corn?
 Verified step by step guidance
Verified step by step guidance1
Step 1: Understand the problem. The goal is to determine the order of the genes (C1, Sh, Wx) in corn based on the phenotypic data provided. This involves identifying the parental, single crossover, and double crossover classes from the data.
Step 2: Identify the parental phenotypes. The parental phenotypes are the most frequent classes in the data. Look for the two phenotypes with the highest numbers, as these represent the original combinations of alleles.
Step 3: Identify the double crossover phenotypes. The double crossover phenotypes are the least frequent classes in the data. These represent the rarest recombination events where two crossovers have occurred.
Step 4: Determine the gene order. Compare the parental phenotypes with the double crossover phenotypes. The gene that differs between the parental and double crossover phenotypes is the middle gene in the sequence.
Step 5: Verify the gene order using single crossover phenotypes. The single crossover phenotypes are intermediate in frequency. Analyze these to confirm the gene order by observing which gene combinations are altered in single crossover events.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
6mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Gene Linkage
Gene linkage refers to the tendency of genes located close to each other on the same chromosome to be inherited together during meiosis. This phenomenon can affect the ratios of phenotypes observed in offspring, as linked genes do not assort independently. Understanding gene linkage is crucial for determining the order of genes based on the frequency of recombination events observed in the offspring.
Recommended video:
Guided course

Chi Square and Linkage
Phenotypic Ratios
Phenotypic ratios represent the relative frequencies of different observable traits in the offspring resulting from a genetic cross. In this case, the ratios of various kernel phenotypes can provide insights into the genetic makeup and linkage of the traits being studied. Analyzing these ratios helps in deducing the arrangement of genes on the chromosome.
Recommended video:
Guided course

Mutations and Phenotypes
Recombination Frequency
Recombination frequency is a measure of how often two genes on the same chromosome are separated during meiosis due to crossing over. It is calculated by dividing the number of recombinant offspring by the total number of offspring. This frequency is used to infer the relative positions of genes on a chromosome, with lower frequencies indicating closer proximity between genes.
Recommended video:
Guided course

Recombination after Single Strand Breaks
Related Videos
Related Practice
Textbook Question
488
views


