Compare DNA transposons and retrotransposons. What properties do they share?
Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
16. Transposable Elements
Transposable Elements in Eukaryotes
Problem 30
Textbook Question
It has been noted that most transposons in humans and other organisms are located in noncoding regions of the genome—regions such as introns, pseudogenes, and stretches of particular types of repetitive DNA. There are several ways to interpret this observation. Describe two possible interpretations. Which interpretation do you favor? Why?
 Verified step by step guidance
Verified step by step guidance1
Step 1: Understand what transposons are—segments of DNA that can move around within the genome, often called 'jumping genes.' Recognize that their presence in noncoding regions means they are found in parts of the DNA that do not code for proteins, such as introns, pseudogenes, and repetitive DNA sequences.
Step 2: Consider the first interpretation: transposons preferentially insert into noncoding regions because these areas are less likely to disrupt essential genes, thus minimizing harmful effects on the organism. This suggests a selective pressure that favors transposon insertions in 'safe' genomic regions.
Step 3: Consider the second interpretation: transposons may insert randomly throughout the genome, but those that land in coding regions are often deleterious and removed by natural selection, leaving a higher observed frequency in noncoding regions over time.
Step 4: Evaluate which interpretation is more plausible by thinking about the mechanisms of transposon insertion and the role of natural selection in shaping genome composition. Reflect on evidence from genetic studies that support either targeted insertion or selection against harmful insertions.
Step 5: Formulate your favored interpretation by weighing the evidence and reasoning. For example, you might favor the idea that natural selection removes harmful insertions in coding regions, leading to an accumulation of transposons in noncoding regions, because this aligns with known evolutionary principles and observed genetic patterns.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
1mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Transposons and Their Genomic Distribution
Transposons are DNA sequences that can move within the genome, often called 'jumping genes.' In many organisms, including humans, they are predominantly found in noncoding regions like introns, pseudogenes, and repetitive DNA. Their distribution affects genome structure and function, influencing gene regulation and genome evolution.
Recommended video:
Guided course

Human Genome Composition
Noncoding Regions of the Genome
Noncoding regions do not code for proteins but include introns, pseudogenes, and repetitive DNA. These areas can regulate gene expression, maintain chromosome structure, or serve as 'safe havens' for transposons, minimizing harmful effects on essential genes. Understanding these regions helps explain why transposons accumulate there.
Recommended video:
Guided course

Genomics Overview
Interpretations of Transposon Localization
Two main interpretations explain transposon localization: (1) natural selection favors transposons in noncoding regions to avoid disrupting vital genes, and (2) transposons preferentially insert into noncoding DNA due to sequence or chromatin preferences. Evaluating these helps understand genome stability and evolutionary pressures.
Recommended video:
Guided course
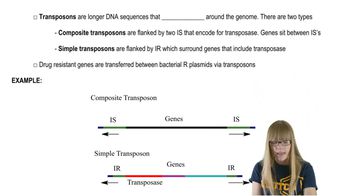
Prokaryotic Transposable Elements

 3:14m
3:14mWatch next
Master Eukaryotic Transposable Elements with a bite sized video explanation from Kylia
Start learningRelated Videos
Related Practice
Textbook Question
1260
views
