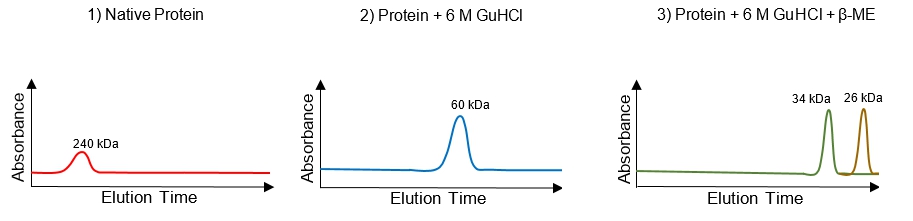
Size exclusion chromatography (SEC), also known as gel filtration chromatography, is a technique used to purify proteins based on their size. The stationary phase in SEC consists of porous beads that create a gel-like environment, allowing for the separation of proteins as they pass through a column. Unlike gel electrophoresis, where smaller molecules migrate faster, in SEC, larger proteins elute first. This is due to the physical size of the proteins in relation to the pores in the beads.
The beads in the stationary phase have cavities of varying sizes, which selectively allow proteins to enter. Large proteins cannot fit into these pores and thus take a shorter route around the beads, leading to their quicker elution. In contrast, smaller proteins can enter the pores, which slows their movement as they navigate the longer pathways within the beads. This results in a distinct elution order: large proteins elute first, followed by medium-sized proteins, and finally, the smallest proteins.
To visualize this process, a chromatogram can be generated, plotting light absorbance against elution time. The chromatogram will show peaks corresponding to the different protein sizes, with the largest proteins appearing first on the left side of the graph, indicating a shorter elution time. This method not only allows for the separation of proteins but can also be used to estimate the size of unknown proteins by comparing their elution times to those of known standards.
Overall, size exclusion chromatography is a valuable technique in biochemistry for purifying proteins and analyzing their sizes, providing insights into their structure and function.