Textbook Question
Compare and contrast introns and exons.
 Verified step by step guidance
Verified step by step guidance Verified video answer for a similar problem:
Verified video answer for a similar problem:
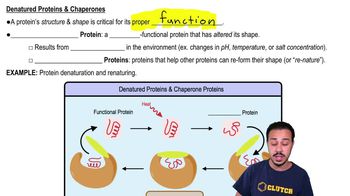


 1:56m
1:56mMaster Eukaryotic Post-Transcriptional Regulation with a bite sized video explanation from Jason
Start learning